SpikePro Help Page
⦿ How to submit a query to SpikePro
The only input that SpikePro requests is the amino acid sequence of the spike protein in
FASTA format.
⦿ SpikePro results and interpretation
Upon submission of the query, SpikePro provides the "Job ID" and a link that must be followed or bookmarked
to check the status of the job. The results are available on the page as soon as the computations are completed
(you may need to refresh the page). They remain on the webserver for two weeks. For each query, SpikePro reports
the following information:
-
The natural logarithm of the predicted fitness of the spike protein sequence submitted by
the user. A value of zero means that it has the same fitness as the original Wuhan spike protein. Negative
fitness values (in green) mean that it is less fit than the Wuhan spike protein, and positive fitness values (in red) that it
is more fit. The predicted fitness is a function of three contributions that describe:
- The virus transmissibility, predicted from the spike protein stability.
- The virus infectivity, predicted from the binding affinity of the spike protein with the ACE2 receptor.
-
The viral escape from the human immune system predicted from the binding affinity of the spike protein
for a series of nAbs.
You can also download the results file generated by SpikePro ( note that the values in this file are not log transformed ! )
For technical details about the method see F.Pucci and M. Rooman, Viruses 2021, 13(5),935
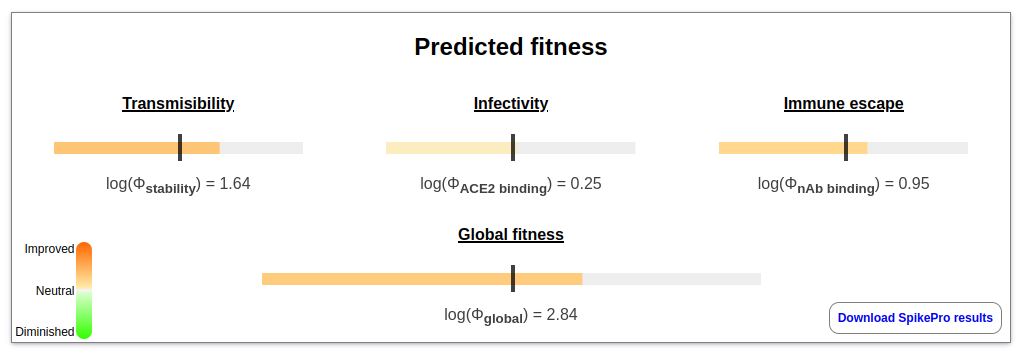
-
The predicted basic reproduction rate R0 of the viral strain computed as a linear function of the
natural logarithm of the predicted fitness
(R0 = 2.7 + 1.39 * log(global fitness)).

-
Predictions based on experimental data (when available) from high-throughput techniques:
-
Viral escape fraction from SARS-CoV-2 infected patient's plasma. Values range from 0 (mutations have no effect on plasma antibody binding)
to 1 (the mutations lead to a complete escape from plasma antibody binding). See Greaney et al., Cell Host Microbe 2021 29, 463-476 for full details.
-
Viral escape fraction from Moderna vaccinated patients. Values range from 0 (mutations have no effect on serum neutralization)
to 1 (the mutations lead to a complete escape from serum neutralization). See Greaney et al., Sci Transl Med 2021 13(600), eabi9915 for full details.
-
Relative change in ACE2 binding constant (log10(KDwuhan) - log10(KDvariant). Positive values
indicate higher binding, while negative values indicate weaker binding between the spike protein and ACE2. See Starr et al., Cell 2020, 182 (5), 1295-1310.e20 for full details.
The values are obtained by adding the experimental values of each individual mutation in the submitted sequence.

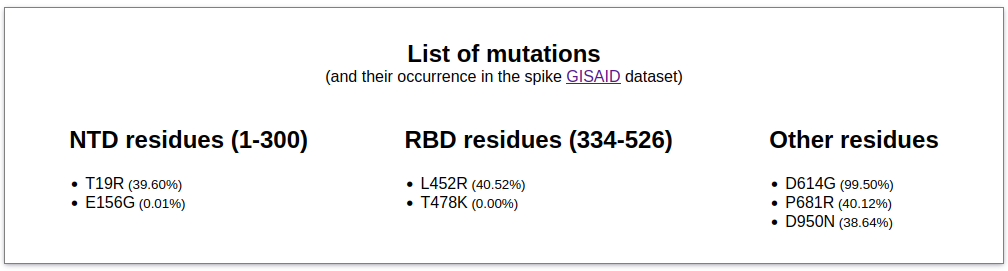
-
A list of the variants in the spike protein sequence submitted by the user with respect to the sequence
deposited in UniProt with the code P0DTC2, which corresponds to the spike protein in the
original Wuhan strain. The localization of the variant residues in the receptor-binding domain (RBD), the
N-terminal domain (NTD) or in other regions of the sequence is also indicated. In addition, the occurrence of each mutation in GISAID's weekly
updated spike protein variants dataset is also reported (note that a registered account on GISAID is required to access their dataset.)
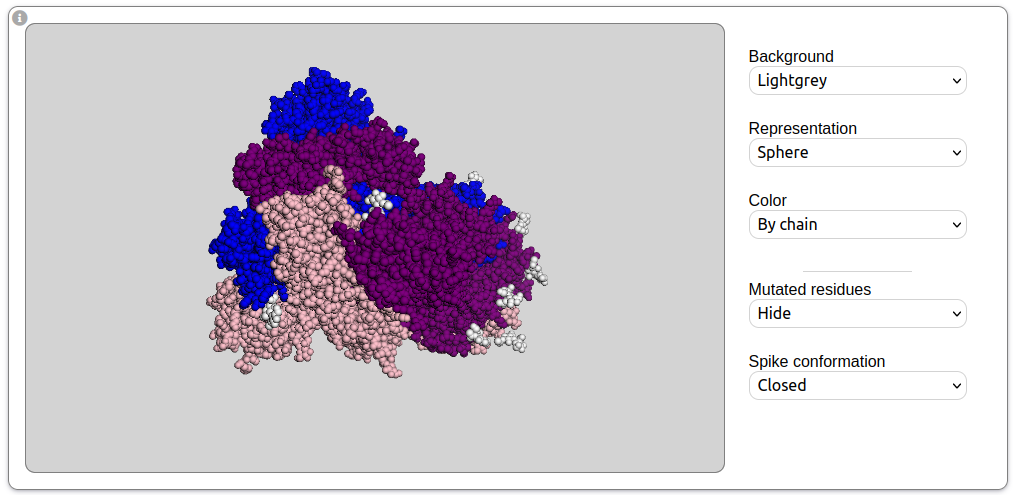
-
A tool to visualize the 3D structure of the complete trimeric spike protein.
Three conformations are available: the closed structure (PDB code: 6VXX), the partially open structure
(PDB code: 6VYB) in which one of the three monomers has its receptor biding domain in an "up" conformation,
and the open structure in which all three monomers are in open conformation (obtained by modeling from the
6VYB structure). The three spike protein chains as well as the mutated residues can be colored using the
side panel buttons. For viewer controls, hover over the information icon on the top left corner.
⦿ Contact
Do not hesitate to contact us at Gabriel.Cia@ulb.be, Jean.Marc.Kwasigroch@ulb.be, Marianne.Rooman@ulb.be or
Fabrizio.Pucci@ulb.be. Feedback is more than welcome.
This webserver has been tested on Firefox, Google-Chrome and Safari.








