The first step is to provide the structure of the protein. You may either provide the 4-letter code of the PDB structure, which will then be retrieved from the Protein Data Bank, or upload your own structure file, which must comply with the PDB format. Please note that: The default structure file on the Protein Data Bank does not necessarily correspond to correct quaternary structure of the protein. Select the extension .pdb1, .pdb2, or .pdb3 to use the corresponding Biological Assembly. The format of the "Biological Assembly" files from the Protein Data Bank is somewhat different from the PDB format. In particular, different chains may be referred to as different models of the same chain. In such cases, SWOTein may have to assign new chain names. In case the structure file contains multiple NMR models, only the first one will be considered.
Once the structure file is uploaded/downloaded, SWOTein provides the user with a summary of its content. For each chain, the name (if available) and number of residues is given.
The second step is to define the chains used for the query. Each protein chain present in the structure file can be selected for the computations, or be discarded. Strengths and weaknesses will not be computed on discarded chains, however they will be taken into account for the solvent accessibility computation of selected chains. 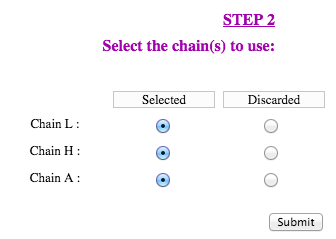
 | Upon completion of the second step and submission of the query, SWOTein provides the "Job ID" and a link that has to be followed/bookmarked to check the status of the job. The results will be available on this page as soon as the computations are over (you may need to refresh the page). They will remain on the webserver for two weeks. For each query, SWOTein reports the following information:A text file containing the ΔG per-residue folding free energies for each residue of the selected chains. Three types of per-residue free energies are displayed accordingly to the different conformational descriptor used in their calculation: "dis" (distance), "tor" (torsion angle domain) and "acc" (solvent accessibility). A table with the ΔG per-residue folding free energies for each residue of the protein colored accordingly to their strength and weakness classification: 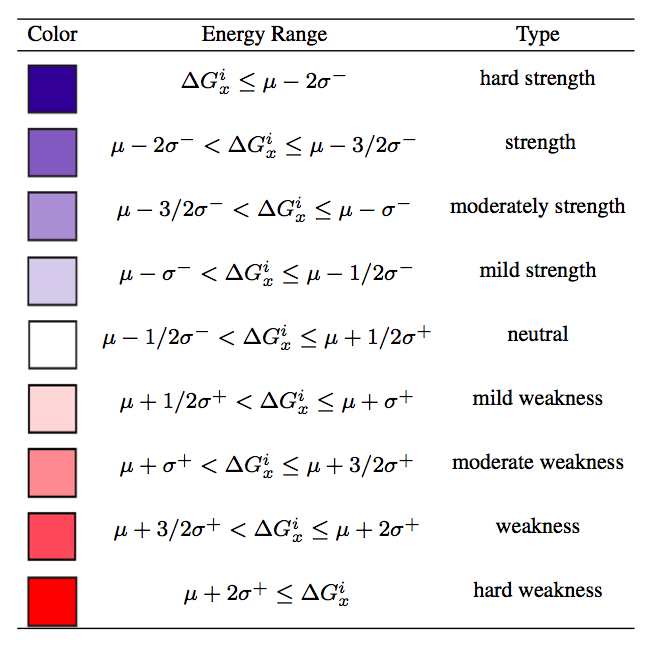
where i indicates the residue, x the type of potential ("dis", "tor" or "acc" ), μ, σ+ and σ- the mean, the right and left standard deviation, respectively, of the corresponding per-residue folding free energy distribution computed on a non-redundant dataset of about 3000 monomeric proteins with high-resolution 3D structures. The values of μ, σ+ and σ for each of the potentials are reported in the following table: 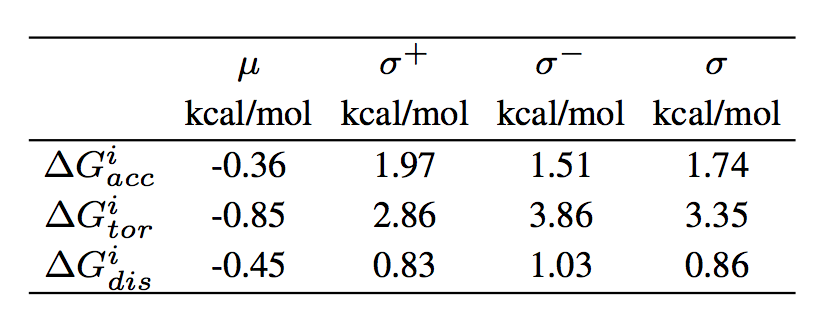
A visualization tool to show the protein 3D structure in which each residue is colored accordingly to residue strenghts/weakness. Three different button are shown on the left of the visualization panel to allow the user to chose the type of potentials used in the visualization. The user can zoom on the residue of interest by clicking on it: all atoms with their positions inclusing the side chain ones will be then shown in that case. Finally we have also implemented a series of additional functionality; it is possible to hide/show a specific chain, select a full screen visualization mode and take a ".png" snapshot of the visualized structures. This page has been tested on Firefox, Google-Chrome, Safari, Edge and Opera. Note that modified residues could not be shown properly.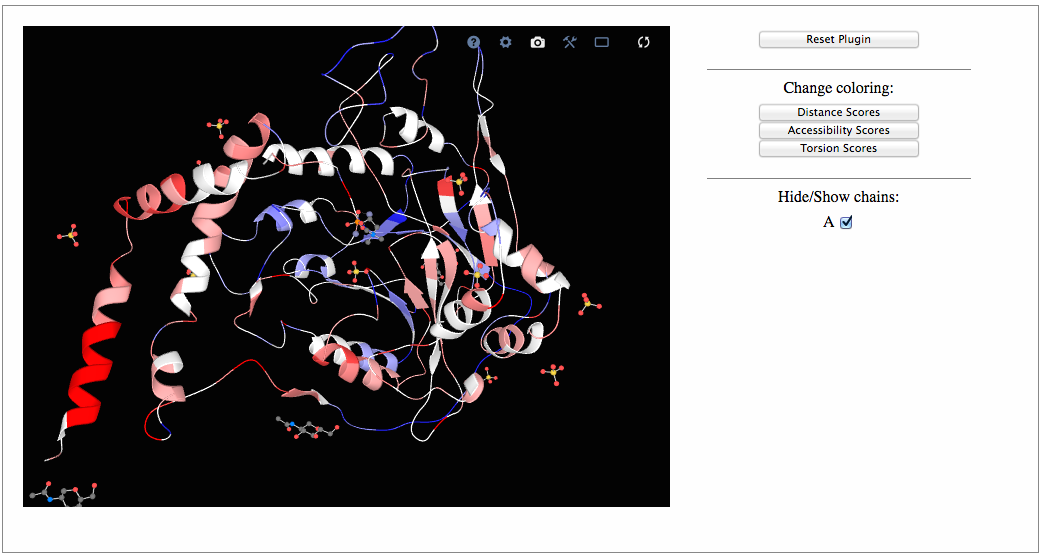
 | The identification of sequence-unique entities relies on the numbering of the amino acids in the structure file. If two chains are identical but numbered differently, they will not be recognized as a sequence-unique entity. In such cases, you may want to edit your structure file and adapt the numbering. This may also be used to introduce a mutation in one chain only and not in other identical chains present in the structure file. In some structure files, chain names are numbers rather than letters. SWOTein does not always deal correctly with this. Try editing your structure file and changing the chain names. SWOTein method is constructed for globular proteins and it is not supposed to work properly for membrane proteins. Be aware that non-protein molecules (RNA, DNA, ions, ligands, etc..) are not taken into account during the calculations. Note that larger errors should be expected for residues in the vicinity of these non-protein molecules.
|  | Do not hesitate to contact us at mrooman@ulb.ac.be. Feedbacks are most than welcome. |
|
|
|