





| |
Upon submission of the query, SOLart provides a "Job ID" and a link that must be followed or bookmarked to check the status of the job. The results are available on this page as soon as the computations are completed (you may need to refresh the page). The results remain on the webserver for two weeks. To predict the solubility of the target protein, SOLart utilizes solubility-dependent statistical potentials, derived from sets of soluble and aggregation-prone proteins. The potentials are of three types : distance potentials that describe tertiary interactions, backbone torsion angle potentials that describe local interactions along the polypeptdide chain and solvent accessibility potentials. SOLart also uses the protein length and amino acid composition of the query proteins as features. The prediction result is reported as a scaled solubility value ranging from 0 to 130. The higher the value, the more soluble the query protein. In addition, SOLart also provides several figures describing the contribution of the top features to the solubility prediction of the target. These features are the "TSS" torsion angle potential, the "ASS" solvent accessibility potential, the "SDS" distance potential, and the protein length (for more details, see the preprint "SOLart: a structure-based method to predict protein solubility"). 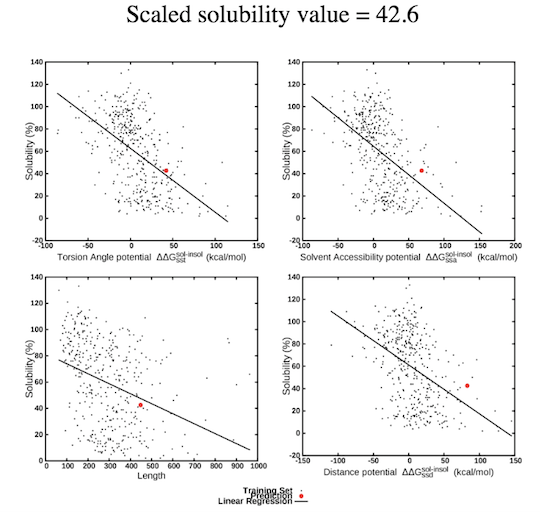 SOLart is very fast and allows a large-scale analyses of the solubility properties of proteins and of the strategies that they use to modulate them.
Our prediction method is designed for globular proteins. Transmembrane proteins are already excluded from the training dataset. Be aware that non-protein molecules (RNA, DNA, ions, ligands, etc..) are not taken into account in the calculations. Thanks for visiting and using SOLart. Do not hesitate to contact us at qinghou@ulb.ac.be, fapucci@ulb.ac.be, jkwasigr@ulb.ac.be or mrooman@ulb.ac.be. Feedback is most welcome. |